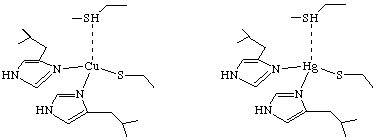
Scheme 1
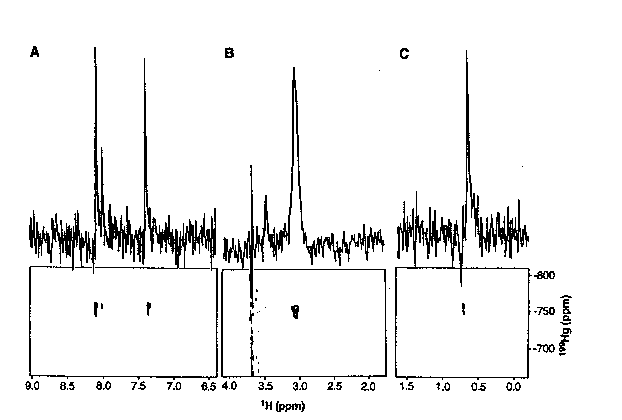
Figure 1
© 1997, Dissolved In Water Incorporated. CEO's: Mike
Lewis, Emma Teuten, and Paul Benny. All rights reserved.
|
|
Project
The study by Utschig, Bryson, and O'Halloran (Utschig, L. M.; Bryson, J. W.; O'Halloran, T. V. Science 1995, 268, 380.) used mercury-199 NMR to determine the structure of the active site for the mercuric ion receptor (MerR) protein, and how this structure might change when the protein is complexed to DNA. MerR is vital for the health of human cells in that it senses changes in the mercury ion concentration, and thus activates the gene responsible for mercury detoxification. Since little is known about mercury-199 NMR, the authors needed to devise their own scale for mercury-199 chemical shifts. This was accomplished by determining the NMR shift of mercury-199 in known coordination geometries.
One of the most common procedure for elucidating the structure of metalloproteins is through the use of cadmium-113 NMR. The authors dismissed this as a possible method for this study because MerR has 288 amino acids, and 1H{113Cd} 2D-NMR experiments generally do not work well on proteins larger than 100 amino acids. Also, previous attempts to produce 1H{113Cd} spectra for MerR yielded no signal. It had previously been suggested that the coordination of mercury in the active site of MerR had at least three cystine side chains bound to the metal when the protein was not complexed to DNA. Therefore, O'Halloran, et. al. wanted to learn if 2D 1H{199Hg} NMR supported these finding, and furthermore, if the active site was affected by complexation to DNA. The first aim of the research was to determine the chemical shifts of mercury in a variety of coordination geometries, and to optimize acquisition parameters. Towards these ends, the authors studied Hg substituted plastocyanin because it had a known structure that was not changed upon substitution of Hg (Scheme 1). The 2D 1H{199Hg} NMR (Figure 1) gave the authors the chemical shift of mercury-199 when it is complexed with two histidines, a cystine, and a methionine in a tetrahedral geometry.


It also gave them optimal delay times for data acquisition. Further studies were required, however, because plastocyanin only has 99 amino acids. Thus, O'Halloran et. al. further tested these optimal delay times on Hg substituted carbonic anhydrase (255 amino acids); a protein of comparable size to MerR. The 2D 1H{199Hg} NMR experiments on carbonic anhydrase determined that the delay times obtained from the plastocyanin studies were sufficient for larger proteins. Therefore, the delay times used for Hg substituted plastocyanin and carbonic anhydrase were used for the 2D 1H{199Hg} NMR study on MerR. Results from the 2D 1H{199Hg} NMR studies show 1H/199Hg coupling that correlate with the coupling of a Beta- C hydrogen from cystine. No similar couplings were obtained that suggest complexation between 199Hg and histidine or methionine. The same 2D 1H{199Hg} spectra was obtained for MerR complexed to DNA. Further evidence for the presence of only cystine amino acids in the DNA complexed MerR was the 1D 199Hg spectra that does not change upon complexation to DNA. Finally, O'Halloran et. al. determined that there was only three cystine residues complexed to the metal center in both the protein, and the DNA complexed protein via the 199Hg shift. The mercury-199 shift of -106ppm (MerR) and -109ppm (MerR-DNA) fall within the established range (-80 to -160ppm) for trisubstituted mercury complexed aliphatic thiolates.
1) Figure 1 shows the 1H{199Hg} 199Hg decoupled spectra for Hg substituted cyanoplastin (Scheme 1). What protons would you assign to peaks 8.07, 7.98, and 7.33? These peaks are examples of _J(1H,199Hg) and _J(1H,199Hg) coupling. (RQD)
2) What would some possible reasons be for the advantage of using 199Hg over the naturally occurring metal in plastocyanin (Cu) and carbonic anhydrase (Zn) for the analysis of active site coordination geometry? (SCL)
3) Is the spectra shown in Figure 1 an example of a heteronuclear 2D J-resolved spectra or a heteronuclear 2D correlated spectra? Explain the difference between the two methods. (SCL)
4) Figure 1 shows a 1H{199Hg} 199Hg decoupled NMR. Is it possible to determine the 1H{199Hg} coupling constants from this spectra? Explain. (ICR)
5) By comparison with known 199Hg four-coordinate mercury compounds, the authors were able to rule out the possibility of MerR being four-coordinate at the active site. Thus, with the knowledge that previous methods had predicted an active site with three cystine residues bound to the metal, they predict a three-coordinate trigonal planar active site in both the protein (MerR) and the DNA complexed protein (DNA-MerR). Is this the only possibility? (EVL)